Scipy stats package#
A variety of functionality for dealing with random numbers can be found in the scipy.stats package.
from scipy import stats
import matplotlib.pyplot as plt
import numpy as np
Distributions#
There are over 100 continuous (univariate) distributions and about 15 discrete distributions provided by scipy
To avoid confusion with a norm in linear algebra we’ll import stats.norm as normal
from scipy.stats import norm as normal
A normal distribution with mean \(\mu\) and variance \(\sigma^2\) has a probability density function \begin{equation} \frac{1}{\sigma \sqrt{2\pi}} e^{-(x-\mu)^2 / 2\sigma^2} \end{equation}
# bounds of the distribution accessed using fields a and b
normal.a, normal.b
(-inf, inf)
# mean and variance
normal.mean(), normal.var()
(0.0, 1.0)
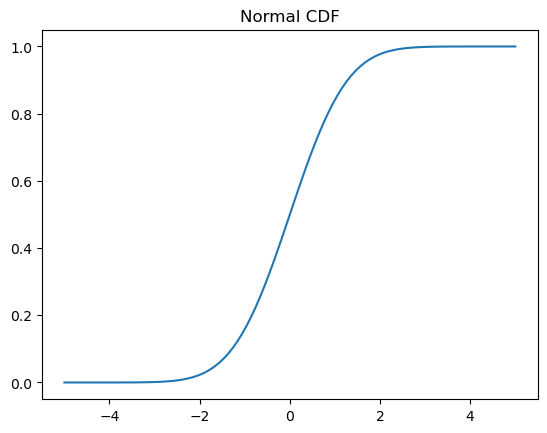
# Cumulative distribution function (CDF)
x = np.linspace(-5,5,100)
y = normal.cdf(x)
plt.plot(x, y)
plt.title('Normal CDF');
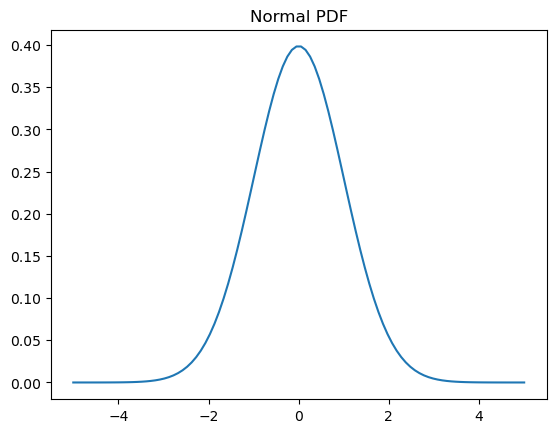
# Probability density function (PDF)
x = np.linspace(-5,5,100)
y = normal.pdf(x)
plt.plot(x, y)
plt.title('Normal PDF');
You can sample from a distribution using the rvs method (random variates)
X = normal.rvs(size=1000)
plt.hist(X);
You can set a global random state using np.random.seed or by passing in a random_state argument to rvs
X = normal.rvs(size=1000, random_state=0)
plt.hist(X);
To shift and scale any distribution, you can use the loc and scale keyword arguments
X = normal.rvs(size=1000, random_state=0, loc=1.0, scale=0.5)
plt.hist(X);
# Probability density function (PDF)
x = np.linspace(-5,5,100)
y = normal.pdf(x, loc=1.0, scale=0.5)
plt.plot(x, y)
plt.title('Normal PDF');
You can also “freeze” a distribution so you don’t need to keep passing in the parameters
dist = normal(loc=1.0, scale=0.5)
X = dist.rvs(size=1000, random_state=0)
plt.hist(X);

Example: The Laplace Distribution#
We’ll review some of the methods we saw above on the Laplace distribution, which has a PDF \begin{equation} \frac{1}{2}e^{-|x|} \end{equation}
from scipy.stats import laplace
x = np.linspace(-5,5,100)
y = laplace.pdf(x)
plt.plot(x, y)
plt.title('Laplace PDF');
x = np.linspace(-5,5,100)
y = laplace.cdf(x)
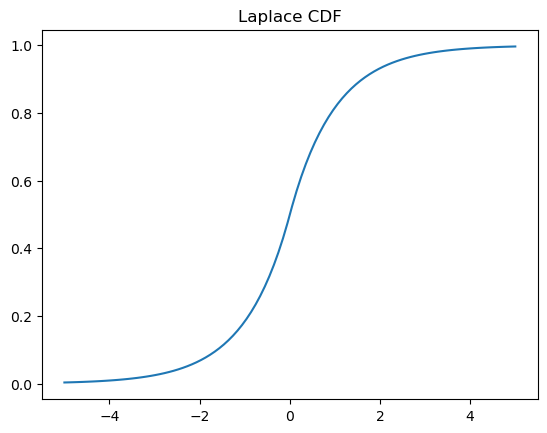
plt.plot(x, y)
plt.title('Laplace CDF');
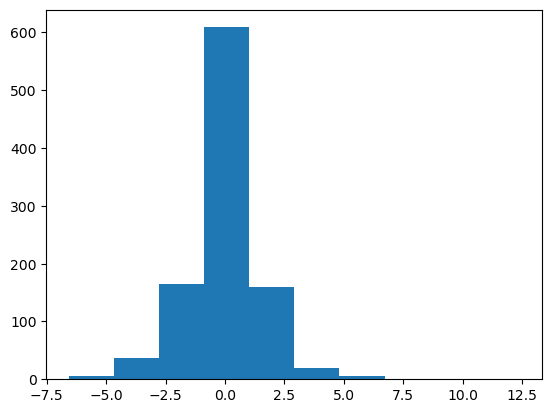
X = laplace.rvs(size=1000)
plt.hist(X);
laplace.a, laplace.b
(-inf, inf)
laplace.mean(), laplace.var()
(0.0, 2.0)
Discrete Distributions#
Discrete distributions have many of the same methods as continuous distributions. One notable difference is that they have a probability mass function (PMF) instead of a PDF.
We’ll look at the Poisson distribution as an example
from scipy.stats import poisson
dist = poisson(1) # parameter 1
dist.a, dist.b
(0, inf)
dist.mean(), dist.var()
(1.0, 1.0)
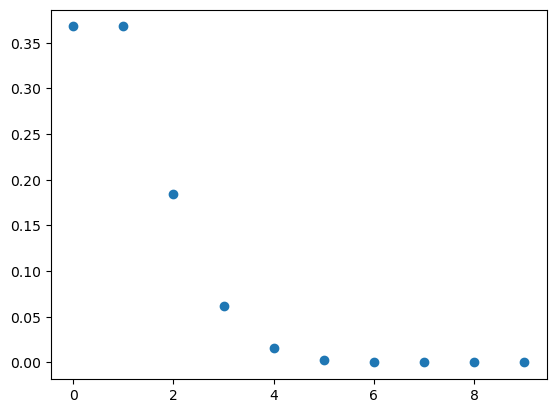
x = np.arange(10)
y = dist.pmf(x)
plt.scatter(x, y);
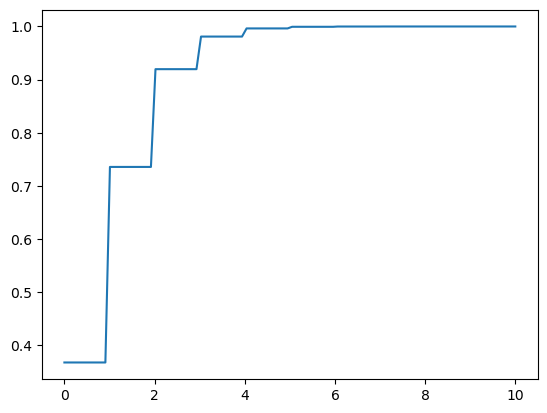
x = np.linspace(0,10, 100)
y = dist.cdf(x)
plt.plot(x, y);
X = dist.rvs(size=1000)
plt.hist(X);
Fitting Parameters#
You can fit distributions using maximum likelihood estimates
X = normal.rvs(size=100, random_state=0)
normal.fit_loc_scale(X)
(0.059808015534485, 1.0078822447165796)
Statistical Tests#
You can also perform a variety of tests. A list of tests available in scipy available can be found here.
the t-test tests whether the mean of a sample differs significantly from the expected mean
stats.ttest_1samp(X, 0)
TtestResult(statistic=0.5904283402851698, pvalue=0.5562489158694675, df=99)
The p-value is 0.56, so we would expect to see a sample that deviates from the expected mean at least this much in about 56% of experiments. If we’re doing a hypothesis test, we wouldn’t consider this significant unless the p-value were much smaller (e.g. 0.05)
If we want to test whether two samples could have come from the same distribution, we can use a Kolmogorov-Smirnov (KS) 2-sample test
X0 = normal.rvs(size=1000, random_state=0, loc=5)
X1 = normal.rvs(size=1000, random_state=2, loc=5)
stats.ks_2samp(X0, X1)
KstestResult(statistic=0.023, pvalue=0.9542189106778983, statistic_location=5.848821448664333, statistic_sign=-1)
We see that the two samples are very likely to come from the same distribution
X0 = normal.rvs(size=1000, random_state=0)
X1 = laplace.rvs(size=1000, random_state=2)
stats.ks_2samp(X0, X1)
KstestResult(statistic=0.056, pvalue=0.08689937254547132, statistic_location=1.5363770542457977, statistic_sign=1)
When we draw from a different distribution, the p-value is much smaller, indicating that the samples are less likely to have been drawn from the same distribution.
Kernel Density Estimation#
We might also want to estimate a continuous PDF from samples, which can be accomplished using a Gaussian kernel density estimator (KDE)
X = np.hstack((laplace.rvs(size=100), normal.rvs(size=100, loc=5)))
plt.hist(X);
A KDE works by creating a function that is the sum of Gaussians centered at each data point. In Scipy this is implemented as an object which can be called like a function
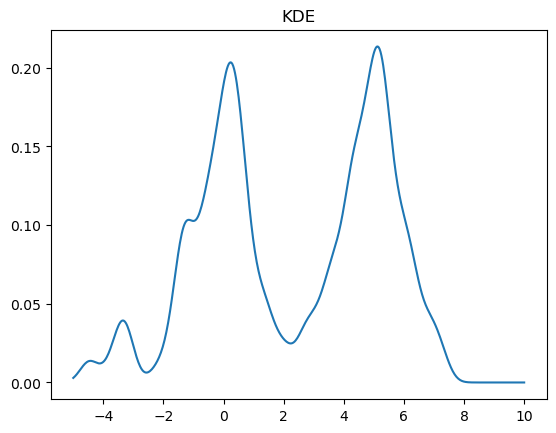
kde = stats.gaussian_kde(X)
x = np.linspace(-5,10,500)
y = kde(x)
plt.plot(x, y)
plt.title("KDE");
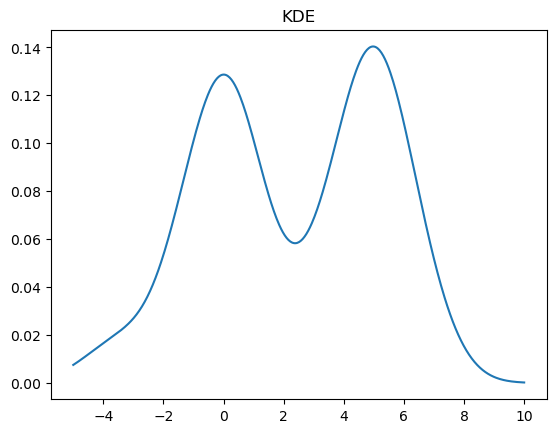
We can change the bandwidth of the Gaussians used in the KDE using the bw_method parameter. You can also pass a function that will set this algorithmically. In the above plot, we see that the peak on the left is a bit smoother than we would expect for the Laplace distribution. We can decrease the bandwidth parameter to make it look sharper:
kde = stats.gaussian_kde(X, bw_method=0.2)
x = np.linspace(-5,10,500)
y = kde(x)
plt.plot(x, y)
plt.title("KDE");
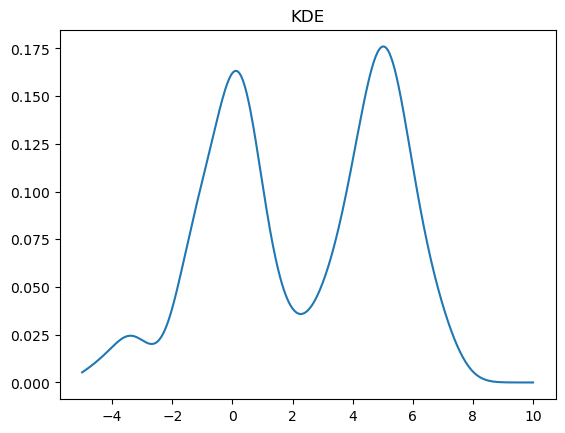
If we set the bandwidth too small, then the KDE will look a bit rough.
kde = stats.gaussian_kde(X, bw_method=0.1)
x = np.linspace(-5,10,500)
y = kde(x)
plt.plot(x, y)
plt.title("KDE");